Phylogeny of series Californicae based on ITS Sequence Data
Goals
To research the evolutionary history of Iris section Limniris series Californicae.
Data
The ITS region is an approximately 630 kb region that includes two spacer regions, ITS1 and ITS2, and the 5.8S ribosomal subunit. The entire aligned data set had 673 characters of which almost 50% were variable.

Analyses
Maximum likelihood where the most likely tree based on the data and chosen model of nucleotide evolution was used. For this analysis I used a transitions (interchanges between pyrimidines, C & T, or between purines, G & A) to transversions (interchanges between pyrimidines and purines) ratio of 1.5.
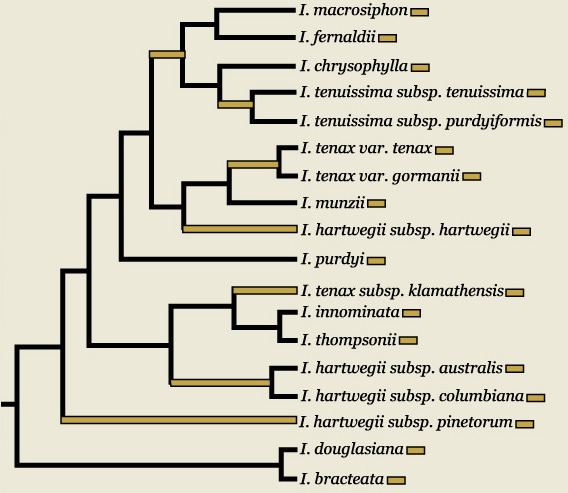
Research from Carol A. Wilson, 2003. Phylogenetic relationships in Iris series Californicae based on ITS sequences of nuclear ribosomal DNA. Systematic Botany 28: 39-46.
Important Findings
Long tubed species
Five of the six taxa with long floral tubes (I. macrosiphon, I. fernaldii, I. chrysophylla, I. tenueissima subsp. tenuissima and I. t. subsp. purdyiformis) were placed together in a monophyletic group (comprised one lineage). Lenz (1958) proposed that these taxa together with I. purdyi, that also has a long floral tube, formed an evolutionary group. My data places I. purdyi outside of this group but does confirm the evolutionary relationships of the other five species.
Iris tenuissima species complex
The Iris tenuissima species complex was resolved as monophyletic. Both Foster (1937) and Lenz (1958) recognized these two taxa as subspecies.
Iris tenax species complex
My data indicates that I. tenax is polyphyletic because I. t. susp. klamathensis was placed in a lineage with I. innominata and I. thompsonii. Lenz (1958) although placing I. t. subsp. klamathensis within I. tenax noted that it had a longer floral tube and longer, narrower style crests than the other two taxa within I. tenax.
Iris hartwegii species complex
Molecular data indicates that the taxa within the I. hartwegii species complex are not monophyletic. I consider this result tentative because the branches leading to I. h. subsp. pinetorum and I. h. subsp. hartwegii were not well supported within my resulting tree. The variability present within the data set provided good resolution between closely related species but was more problematic along the backbone of the tree.
G-C content of sequences
The G-C content of the sequences ranged from 57 to 69%, a percentage that is unusually high within angiosperms. Such high G-C content increased the difficulty of sequencing these taxa and resulted in a few unresolved areas in some nucleotide sequences. As an example, in I. hartwegii (G-C content = 65%) there were 10 unresolved nucleotides sites that were adjacent to a long string of C’s.
Conclusions
Of the three species complexes within Iris series Californicae, only I. tenuissima was resolved as monophyletic. Iris tenax subsp. klamathensis was not placed with other members of the I. tenax species complex. The four members of the I. hartwegii complex were placed in three separate lineages although this result is considered tentative awaiting further study. The ITS region was highly variable for this group and resolved relationships between closely related species. The high variation in the data set presented problems of homoplasy in nucleotide characters that confounded relationships along the backbone of the tree. The high G-C content of many sequences increased the difficulty of obtaining data.
Acknowledgements
The American Iris Society and the Society of the Pacific coast Native Iris provided funding for this project.
References
Lenz, L. W. 1958. A revision of the Pacific Coast Irises. Aliso 4; 1-72.
Citation
Refer to Wilson, C. A. 2003. A phylogenetic analysis of Iris series Californicae based on ITS sequences of nuclear ribosomal DNA. Systematic Botany 28: 39–46.
